- Files
- Control Panel
- Move & Rotate
- Display
- Rendering
- Tools
- Mutations
- Torsions
- Preferences
- hardware stereo
- electrostatics
- surface
- tips & tricks
Index
by N.Guex
& T.Schwede
Mutating Amino-Acids
Mutating Amino-Acids
Swiss-PdbViewer allows to browse a rotamer library in order to change amino acids sidechains. This can be very useful to quickly evaluate the putative effect of a mutaion before actually doing the lab work.
To initiate a mutation, simply click on the mutation tool located at
the top of the display window. You will be prompted to select a group,
which is readily made by clicking on any atom of the desired amino acid.
Then a pop-up menu will let you select the new amino acid type.
Note: clicking outside the list, as well as hitting the
"return" or "enter" key will automatically select the rotamer library
of the current amino acid.
At this point, the current sidechain will be replaced by the "best" rotamer
of the new amino acid, and the mutation tool will appear in inverse video
to signify that a mutation is under process.
The "best" rotamer is the one that totalize the lowest score
according to the formula:
score = (4 x NbClash with backbone N CA and C atoms) +
(3 x NbClash with backbone O atoms) +
(2 x NbClash with sidechains atoms) -
NbHbonds - 4 x Nb SSbonds
The current rotamer number is displayed just below the tools in the Display Window. You may obtain something like:
rotamer: 4/16 score: -1
which signifies that rotamer 4 out of 16 available has been choosen. This display is updated each time you change the rotamer by clicking on one of the little arrows that come below the mutate tool, or by hitting the "*" key of the numerical keyboard. Note that holding the "shift" key while hitting the "*" key will select the previous rotamer instead of the next one.
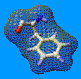
Putative H-bonds will appear in green, and steric hindrances in purple.
Note that they will be drawn only if the group that makes the contact
is visible, so think to enlarge the view around the group to mutate before
proceeding (or even after see below)
You can freely rotate, move and resize the molecule, as well as measure
distances, display, undisplay, groups and so on...
Once you're done with your experimentations, you can end the "mutation"
process by clicking once again on the mutation tool. You will be prompted
for accepting or discarding the mutation (which will put back the initial
group sidechain).